Quickstart#
Run the analysis using the Jupyter notebook at notebook/workflow_2D.ipynb.
Required Files#
Neural data directory (e.g., minian/ output):
{trace_name}.zarr: calcium traces (e.g.,C.zarrorC_lp.zarr)A.zarr: spatial footprints for cell overlay (optional)max_proj.zarr: max projection image for visualization (optional)
Timestamp and position files:
neural_timestamp.csv: neural frame timestampsbehavior_position.csv: animal position with bodypart columns (DeepLabCut format)behavior_timestamp.csv: behavior frame timestamps
Configuration files:
config.yaml: analysis parametersdata_paths.yaml: paths to your data files
Setup#
1. Create data paths config#
Create data_paths.yaml with paths relative to this file:
data_paths.yaml
id: your_data
mio_model: placecell.config.DataPathsConfig
mio_version: 0.8.1
# Directory containing zarr files (C.zarr, A.zarr, max_proj.zarr)
neural_path: path/to/minian_output
neural_timestamp: path/to/neural_timestamp.csv
behavior_position: path/to/behavior_position.csv
behavior_timestamp: path/to/behavior_timestamp.csv
2. Create analysis config#
Create config.yaml with analysis parameters:
config.yaml
id: your_config
mio_model: placecell.config.AnalysisConfig
mio_version: 0.8.1
neural:
id: neural
fps: 20.0
trace_name: C
oasis:
id: oasis
g: [1.60, -0.63]
baseline: p10
penalty: 0
behavior:
id: behavior
behavior_fps: 20.0
bodypart: LED
speed_threshold: 10.0
speed_window_frames: 5
spatial_map_2d:
id: spatial_map_2d
bins: 50
min_occupancy: 0.05
occupancy_sigma: 3
activity_sigma: 3
n_shuffles: 500
p_value_threshold: 0.05
3. Run the notebook#
Open notebook/workflow_2D.ipynb in Jupyter Lab, set CONFIG_PATH and DATA_PATH to your config files, and run all cells.
Output#
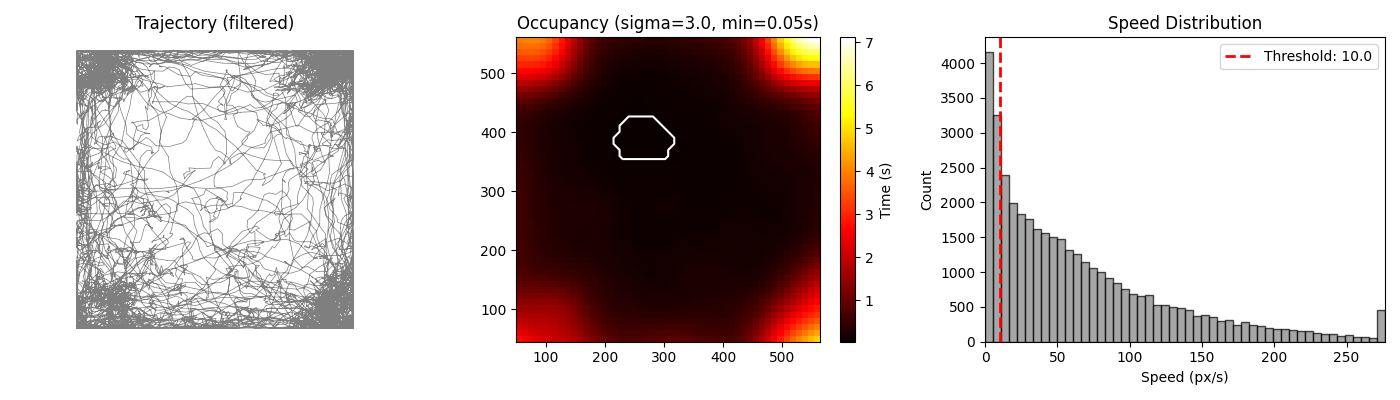
The workflow displays an occupancy preview:
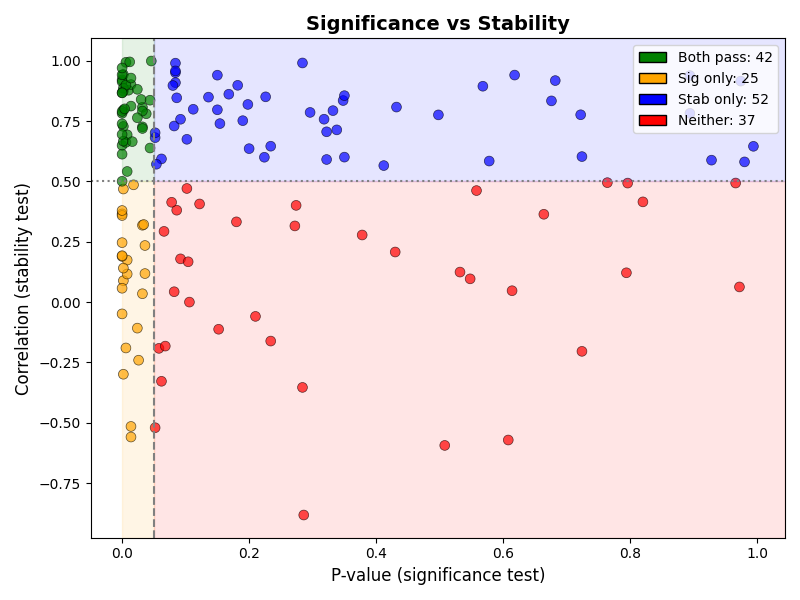
Then displays the stability vs significance plot:
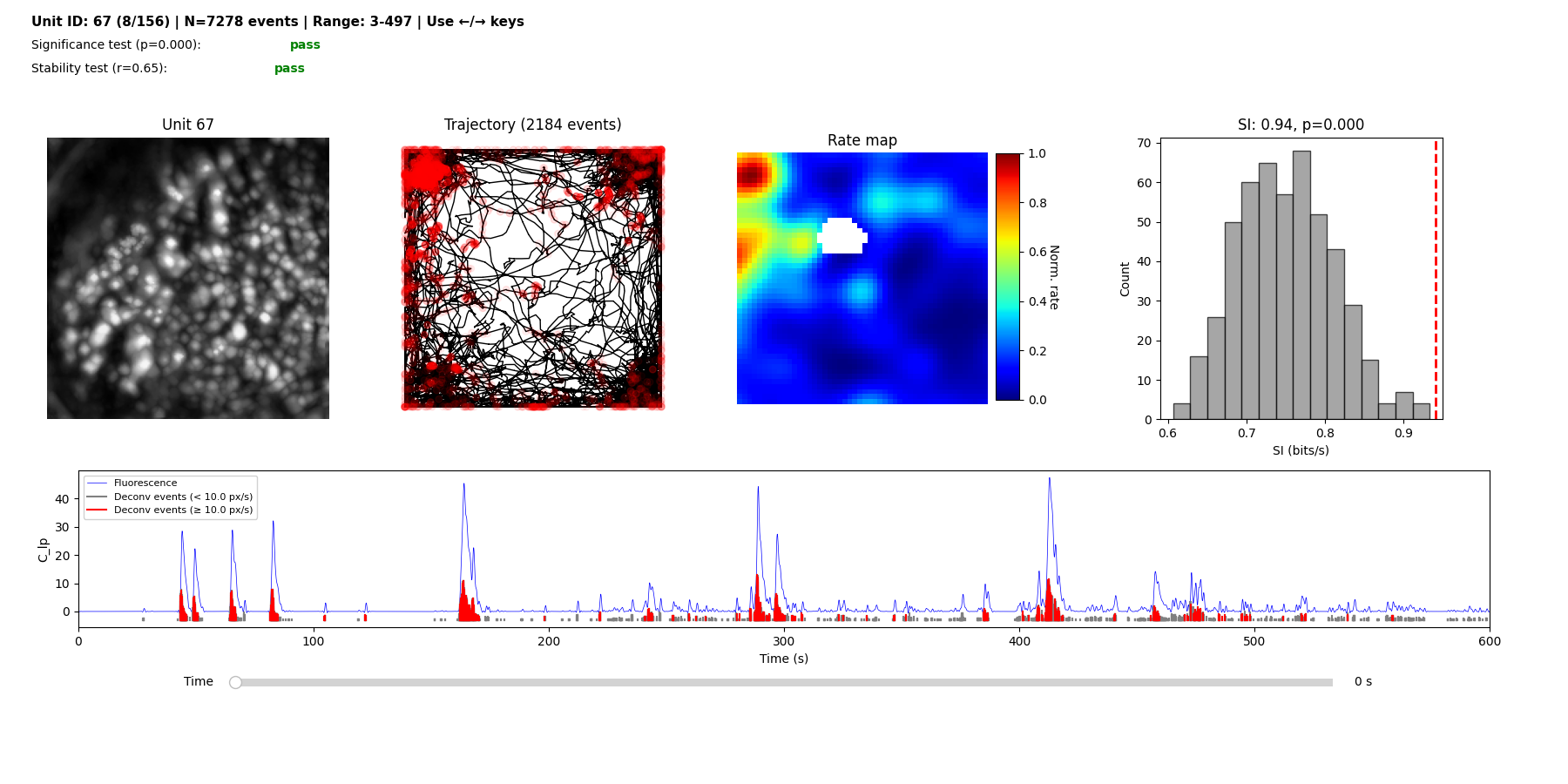
And finally launches the interactive place cell viewer:
Next Steps#
See Pipeline Details for how the analysis works